CSC 458 - Data
Mining & Predictive Analytics I, Spring 2024, Assignment 2, Regression.
Due by 11:59 PM on Thursday February 29 via D2L. We will have
some work time in class.
You will turn in 2 files by the deadline, with a 10% per day
penalty and 0 points after
I go over my solution: CSC458Assn2Small.arff
and README.txt
with your
answers.
Their creation is given in the steps below. I prefer you
turn in a .zip folder (no .7z)
containing only those files. You can turn individual files
if you don't have a zip utility.
As with all assignments except Python Assignment 4,
this is a mixture of two things.
1. Analysis using stable Weka
version 3.8.x. Use this free stable version, not a vendor
version.
2. Answering questions in README.txt
If you are running on a campus PC with the S:\ network drive
mounted, clicking:
s:\ComputerScience\WEKA\WekaWith2GBcampus.bat
starts Weka 3.8.6. Save your work on a thumb
drive or other persistent drive.
Campus PCs erase what you save on their drives
when you log off.
Many students download Weka 3.8.x and work on
their own PCs or laptops.
We are using only 10-fold cross-validation testing in this
assignment for
simplicity.
Assignment Background
Here is CSC458S24RegressAssn2Handout.zip
containing three files:
CSC458S24ClassifyAssn1Handout.arff is
the Weka Attribute Relation File Format
dataset with the starting
data for Assignment 1. We will use this data briefly.
CSC458S24RegressAssn2Handout.arff is
our related dataset for regression
analysis in Assignment 2.
Regression attempts to predict numeric target
attribute values in each
instance based on non-target attributes that may be
numeric or nominal.
README.txt contains questions that you
must answer between some steps.
extractAudioFreqARFF11Feb2024.py is an
updated version of
extractAudioFreqARFF17Oct2023.py used in Assignment 1 for
extracting
data from .wav files.
Please refer to the Assignment
1 handout for background on this audio analysis
project. In Assignment 2 we are regressing
values for the tagged toscgn signal
gain attribute, with values in the range
[0.5, 0.9] on a scale of [0.0, 1.0]
This attribute is present in both of the
handout ARFF files, but we did not use it
in Assignment 1. It is the only tagged
attribute being used in Assignment 2.
A previous
semester's handout, recently updated using SciPy's wav
file reader and
fft frequency-domain histogram extraction,
serves as a reference.
The main difference between Assignment 1's CSC458S24ClassifyAssn1Handout.arff
dataset and Assignment 2's CSC458S24RegressAssn2Handout.arff
dataset is that,
whereas the former normalized ampl2 through
ampl32 as a fraction of the fundamental
ampl1 scaled to 1.0, and normalized freq2
through freq32 as a multiple of the
fundamental frequency of scaled to 1.0, CSC458S24RegressAssn2Handout.arff
contains neither of these scalings. The
amplitude and frequency attributes are
the values extracted by the wav
file read function and the fft
frequency-histogram function.
Figure 1 illustrates that extractAudioFreqARFF11Feb2024.py
has extracted the
fundamental frequency ampl1 as generated by the
line "freq = random.randint(100,2000)"
in the
signal generator, with the off-by-1 difference likely due to
rounding.
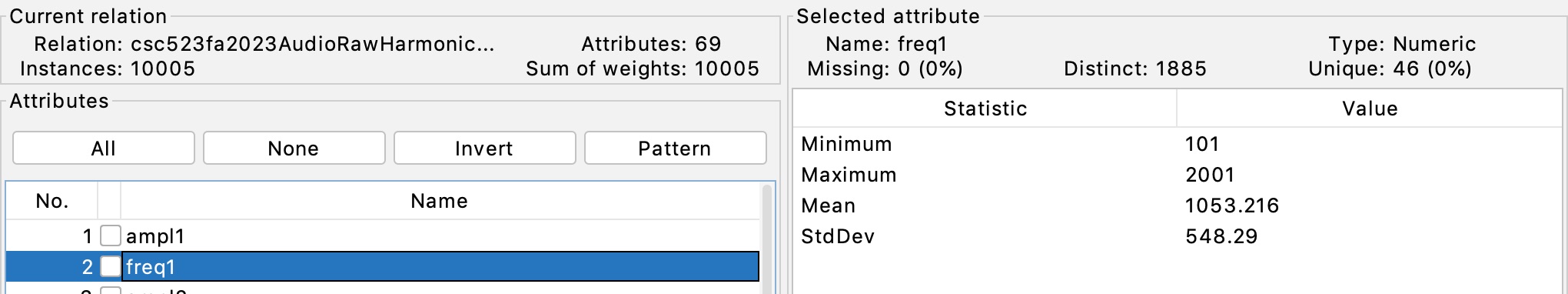
Figure 1: Fundamental frequency ampl1 in range [101.0, 2001.0]
per the
signal generator
STEP 1: Load CSC458S24RegressAssn2Handout.arff
into Weka and Remove tagged attributes
toosc, tfreq, tnoign
and tid as defined in Assignment 1. We will predict signal
gain toscgn in the
range [0.5, 0.9] on a scale of [0.0, 1.0] from
non-tagged, non-target attributes. There should be
65 attributes at this point including toscgn.
NOTE: You can save work-in-progress ARFF files at any time
you take a break or want a backup
of your edits.
STEP 2: In Weka's Classify tab run rules -> ZeroR.
README Q1: Paste this part of ZeroR's result in Q1:
ZeroR predicts class value: N.n
...
Correlation
coefficient
N.n
Mean absolute
error
N.n
Root mean squared
error
N.n
Relative absolute
error
N.n %
Root relative squared
error
N.n %
Total Number of
Instances
10005
README Q2: How does ZeroR arrive at the toscgn
value "ZeroR predicts
class value: N.n"?
Use the Weka Preprocess tab to look for your
answer. Clicking the ZeroR configuration
entry line and then More is also helpful.
README Q3: How good is the correlation coefficient of Q1 in
predicting toscgn?
See Evaluating numeric prediction links under
Assignment 2 on the course page
for evaluating testing results.
STEP 3: In Weka's Select Attributes tab run the
default Attribute Evaluator of
"CfsSubsetEval -P 1 -E 1" with Search Method set to "BestFirst -D
1 -N5" and
hit Start.
README Q4: Paste this subset of the evaluator's output.
Selected attributes: List and number of attribute indices.
... (paste all lines below "Selected attributes")
README Q5: Given what you know about the fundamental
frequency being the
peak measurement on the left side of the
frequency histogram, and the decay rates
of subsequent ampl measures from Assignment
1 Figure 13, why do you think the
ampl attributes uncovered in Q4 appear as
important for prediction of toscgn?
Address ampl attributes appearing in
Q4.
STEP 4: In Weka's Select Attributes tab select the
Attribute Evaluator
"CorrelationAttributeEval", click "yes" for the pop-up Search
Method of
Ranker -- it is ranking attribute correlations to toscgn
-- and hit Start.
README Q6: Paste the top five results outlined here. How do
the first five non-target
attributes pasted in your answer relate to the ranking of STEP 3?
Attribute Evaluator (supervised, Class (numeric): 65 toscgn):
Correlation Ranking Filter
Ranked attributes:
N.n n attributeName
N.n n attributeName
N.n n attributeName
N.n n attributeName
N.n n attributeName
NOTE: The N.n column above is the correlation coefficient
of the non-target
attribute to toscgn, the middle n column is the
attribute number of this non-target
attribute, followed by its name.
README Q7: In Weka's Classify tab run functions ->
SimpleLinearRegression
and paste these output lines. SimpleLinearRegression is the regression counterpart
to classification's OneR, selecting the most strongly correlated
non-target attribute
to correlate with toscgn. What attribute does it select,
does that selection agree with
the most highly correlated attribute in Q4 and Q6, and has the
correlation coefficient
improved over ZeroR?
Linear
regression on attributeName
...
n * attributName + N.n
...
Correlation
coefficient
N.n
Mean absolute
error
N.n
Root mean squared
error
N.n
Relative absolute
error
N.n %
Root relative squared
error
N.n %
Total Number of
Instances
10005
README Q8: In Weka's Classify tab run functions ->
LinearRegression
that attempts to use all correlated attributes. Paste this part
of its result.
Correlation
coefficient
N.n
Mean absolute
error
N.n
Root mean squared
error
N.n
Relative absolute
error
N.n %
Root relative squared
error
N.n %
Total Number of
Instances
10005
Has the correlation coefficient improved over
SimpleLinearRegression?
What do you notice about the multipliers in its linear formula?
(Do not paste this formula, just inspect it.)
toscgn =
n *
attributeName +
n *
attributeName +
README Q9: In Weka's Classify tab run trees -> M5P
that attempts to use all correlated attributes. The M5P decision
tree divides non-linear data relationships into leaves that
are approximations of linear relationships in the form of
linear expressions. Has the correlation coefficient improved
over
LinearRegression? How good is it?
What do you notice about the multipliers in its linear formulas?
What do you notice about the magnitude of values in its decision
tree?
Paste this part of its result.
Correlation
coefficient
N.n
Mean absolute
error
N.n
Root mean squared
error
N.n
Relative absolute
error
N.n %
Root relative squared
error
N.n %
Total Number of
Instances
10005
NOTE: There are two problems with this dataset.
First, the magnitude of the
correlated non-target attribute values are very high, leading to
the apparent
multipliers in the above linear expressions. Second, there are too
many attributes.
STEP 5: Remove all attributes except the top 2 "Selected
attributes" for Q4
plus toscgn. There are only 3 attributes now.
README Q10: In Weka's Classify tab run trees -> M5P and
paste
the following out. How does CC compare to that of Q9?
What do you notice about the multipliers in its linear formulas?
Paste this part of
its result.
Correlation
coefficient
N.n
Mean absolute
error
N.n
Root mean squared
error
N.n
Relative absolute
error
N.n %
Root relative squared
error
N.n %
Total Number of
Instances
10005
STEP 6: In Weka's Preprocess tab run Filter ->
unsupervised -> attribute -> MathExpression
with the default formula "(A-MIN)/(MAX-MIN)". Unlike
AddExpression which creates
a new named derived attribute, MathExpression with default
settings applies its formula
to every non-target attribute in the dataset. Unlike
Assignment 1 where amplitudes
and frequencies were normalized against the amplitude and frequency of the
fundamental frequency, "(A-MIN)/(MAX-MIN)"
normalizes each non-target attribute
as a fraction of its respective
distance between that attribute's
individual minimum and
maximum values. This is a
within-attribute, across-all-instances
normalization, not a
cross-attribute as in Assignment 1's
dataset.
Make sure to Apply and inspect
in Preprocess that the two non-target
attributes have
been mapped, linearly, into the range
[0.0, 1.0], and that toscgn
remains in the range
[0.5, 0.9]. We want predictions in
toscgn application terms.
(SIDE NOTES: 1. After
prepping and reviewing this
assignment, I found that
SciPy's
fft function has a parameter called
norm that when changed
from the default
"backward" to "forward" value, brings
the range of histogram values down
into a lower
range that makes linear expression
multipliers more visible in Weka
without affecting
accuracy of predictions. 2.
The default MathExpression formula
above gives identical
results to Weka's Normalize attribute
filter. 3. In some
assignments we use the
Normalize filter, not to make
multipliers more visible in Weka, but
to put them
on the same within-attribute [0.0,
1.0] range so we can compare their
multipliers
for improtance. 4. I
decided to leave the initial fft
result in place in order to support
practice
using a Weka filter.)
SAVE this 3-attribute dataset
after applying MathExpression
as CSC458Assn2Small.arff
and turn it into D2L with
your README.txt file by
the deadline.
README Q11: In Weka's
Classify tab run trees -> M5P and
paste
the following output. How do CC,
MAE, and RMSE compare to those of
Q10?
What do you notice about the
multipliers in its linear formulas?
Also, how do the MAE (mean absolute
error) and RMSE (root mean squared
error)
measures compare to the toscgn
range of values [0.5, 0.9] and its
mean?
Paste this part of its
result.
Number of Rules : nn
...
Correlation
coefficient
N.n
Mean absolute
error
N.n
Root mean squared
error
N.n
Relative absolute
error
N.n %
Root relative squared
error
N.n %
Total Number of
Instances
10005
README Q12: In
Weka's
Classify tab
select rules
->
DecisionTable,
go into its
configuration
settings and
set displayRules
to true,
and run it.
How many rows
of rules are
there, and how
many linear
formulas
("Number of
Rules) in M5P
for Q11?
How do the
complexity of
Q11's and
Q12's models
compare? How
do their CCs
compare?
Paste
this part of
its result.
Number of
Rules : nn
...
Correlation
coefficient
N.n
Mean absolute
error
N.n
Root mean
squared
error
N.n
Relative
absolute
error
N.n %
Root relative
squared
error
N.n %
Total Number
of
Instances
10005
Number of
training
instances:
10005
README
Q13: In
Weka's
Classify tab select
trees ->
M5P,
go into its
configuration
settings and set
minNumInstances
to
1000.
This increase
will put more
instances at
each leaf than
in the default
value for
minNumInstances,
resulting in
more
shallow trees
that are
possibly
easier to
read.
How many
linear
formulas
("Number of
Rules) in M5P
for Q13
compared to
Q11?
How do the
complexity of
Q11's and
Q13's models
compare?
How do their
CCs compare?
Paste this
part of its
result.
Number of
Rules : n
...
Correlation
coefficient
N.n
Mean absolute
error
N.n
Root mean
squared
error
N.n
Relative
absolute
error
N.n %
Root relative
squared
error
N.n %
Total Number
of
Instances
10005
README Q14:
Load
Assignment 1's
CSC458S24ClassifyAssn1Handout.arff
into Weka,
remove remove
tagged
attributes
toosc, tfreq,
tnoign
and tid.
Run M5P with
the
minNumInstances
parameter set
to the
default value
of 4.
Next remove
all attributes
except the top 2 "Selected attributes" for Q4 plus toscgn.
There are only 3 attributes now.
Run M5P again. What are their CC values? How do these CCs of M5P
using
CSC458S24ClassifyAssn1Handout.arff
compare to the earlier M5P runs with
CSC458S24RegressAssn2Handout.arff
and
CSC458Assn2Small.arff?
/
Why are these CSC458S24ClassifyAssn1Handout.arff
results so substantially different?
Think about the differences between CSC458S24ClassifyAssn1Handout.arff's
versus
CSC458S24RegressAssn2Handout.arff's
values for non-target attributes.
Also, look at the attributes in Q14's decision trees compared to
the earlier
M5P trees using CSC458S24RegressAssn2Handout.arff's data.
Correlation
coefficient
N.n (for the 65-attribute dataset after removing
the other tagged attributes)
Correlation
coefficient
N.n (for the 3-attribute dataset after
removing all but 3 attributes))
README Q15: These are the remaining 6.6% for turning in a
correct CSC458Assn2Small.arff
file.
