CSC 558 - Predictive
Analytics II, Spring 2023, Assignment 3 on time series.
Assignment 3 due by 11:59 PM on Sunday April 9 via D2L
Assignment 3
Answer to a March 29 question added here on March 30:
These
questions came up after most students left Zoom. For SS_All
using the monthly data my Q2 for M5P looks like this (These
are in the last 15 minutes of the zoom recording):
HMtempC_median <=
6.367 : LM1 (71/4.072%)
HMtempC_median > 6.367 :
| HMtempC_mean <=
21.362 :
| | HMtempC_mean <= 16.646 :
| | | HMtempC_median <= 8.858
:
| | | | wndNW <=
92.5 : LM2 (15/4.669%)
| | | | wndNW >
92.5 :
| | | | |
HMtempC_mean_td <= 0.933 : LM3 (2/3.209%)
| | | | | HMtempC_mean_td >
0.933 : LM4 (3/1.963%)
| | | HMtempC_median >
8.858 : LM5 (47/100.344%)
| | HMtempC_mean > 16.646 : LM6
(52/34.273%)
| HMtempC_mean > 21.362 : LM7
(39/1.432%)
Correlation
coefficient
0.699
Mean absolute error
688.5367
Root mean squared error
1240.3141
Relative absolute error
52.7569 %
Root relative squared error
71.2516 %
Total Number of Instances
229
To
figure out importance of the attributes in the decision tree,
the ones closest to the left are the most
important. They are near the root and
split the decision subspace below them, ideally in half. For
the above tree, the most important attributes are:
HMtempC_median
(0 steps from left)
HMtempC_mean
wndNW
HMtempC_mean_td
There
are only 4 in the above tree. Also, if you get a tie with
respect to depth in the tree (from the left), just list them
both.
For
LinearRegression on NORMALIZED non-target attributes, just
look at the absolute values of the multipliers. The top 5 are
in bold below.
SS_All
=
-60.2765 * WindSpd_mean +
85.1711 * HMtempC_median +
94.0799 * WindSpd_median +
1042.8756 * HMtempC_pstdv +
159.5224 * WindSpd_pstdv +
223.4029 * HMtempC_min +
96.506 * WindSpd_min +
-305.1059 * HMtempC_max +
-33.1143 * WindSpd_max +
15.1031 * wndN +
-90.3262 * wndNNE +
51.6743 * wndE +
50.7002 * wndSW +
34.8321 * wndW +
23.2225 * wndNW +
-512.5064 * HMtempC_pstdv_td +
-110.5418 * HMtempC_min_td +
155.4989 * HMtempC_max_td +
-30.3891 * wndE_td +
-23.1375 * wndSW_td +
-19.7927 * wndW_td +
-9.5087 * wndNW_td +
-3033.4981
Time
taken to build model: 0.01 seconds
===
Cross-validation ===
=== Summary ===
Correlation
coefficient
0.5114
Mean absolute error
1140.164
Root mean squared error
1537.7767
Relative absolute error
87.3614 %
Root relative squared error
88.3398 %
Total Number of Instances
229
For
Q19 it'll just be the bottom lines of the result from Select
Attributes tab:
Selected
attributes: 5,11,15,19,21,23,25,28 : 8
HMtempC_pstdv
wndN
wndE
wndS
wndSW
wndW
wndNW
HMtempC_mean_td
If
you need only 5 of those attributes in a question, just use
the first 5 listed.
This is an extension of an ongoing
study to correlate climate change to declines in raptor
counts at Hawk Mountain Sanctuary.
Here are slides
from last fall's seminar. Here is a CSC458
assignment using log10 compression of raptor
counts.
Download compressed ARFF data files week_change.arff.gz
and month_change.arff.gz
Q&A file README.assn3.txt
from these links.
You must edit an ARFF file and answer questions in
README.assn3.txt and turn it in to D2L by the deadline.
This is a crowd-sourcing assignment that is a preliminary new
study to search for data patterns. It is not cooked to
convergence.
There is a 10% late penalty for each day the assignment is late.
STEP 0: A RandomTree of student assignments.
Each student must select a raptor species from 1 of 8 to analyze
using the following approach.
Take a coin and toss it without cheat three times. Assign it
left-to-right according to the following table.
$ python raptors.py
Tail
Tail Tail
AK_All American Kestrel
Tail Tail
Head BW_All Broad-wing Hawk
Tail Head
Tail CH_All Cooper's Hawk
Tail Head
Head NG_All Northern Goshawk
Head Tail
Tail NH_All Norther Harrier
Head Tail
Head OS_All Osprey
Head Head
Tail RL_All Rough-legged
Hawk
Head Head
Head RT_All Red-Tail Hawk
This script generates that table.
$ cat raptors.py
coin = ['Tail', 'Head']
toss = []
raptors = [ 'AK_All', 'BW_All', 'CH_All', 'NG_All', 'NH_All',
'OS_All', 'RL_All', 'RT_All']
index = 0
for c1 in coin:
for c2 in coin:
for c3 in coin:
print(c1 + '\t' + c2 + '\t' + c3 + '\t' + raptors[index])
index += 1
Your raptor to analyze is selected by that table. Keep track of
its two-letter symbol.
I have taken SS (Sharp-shinner Hawk) and PG (Peregin Falcon) for
myself.
I must keep 3 related target attributes in my dataset, usinmg one
at a time.
XX_All
where XX is SS for me. This
is the count of SS during that time period.
XX_All_log10 is the
base 10 logarithm used for target raptor count compression. We
will discuss on March 22.
XX_All_td
is the time-series difference from the previous
year's measure during that time band (week or month).
_td looks to see whether there is
a consistent change during a given month or week across
consecutive years.
week_change.arff_td.csv.gz
and month_change.arff_td.csv.gz
are sorted tables by year for each _td time series.
This
zip directory gives all files used in preparing this
assignment. I will go over them March 22 and 29.
State your name and XX raptor selection at the
top of README.assn3.txt.
STEP 1: Preprocessing / Filtering
1a. Load
month_change.arff.gz into Weka.
1b. Create a Filter -> unsupervised -> attribute ->
AddExpression that creates wnd_WNW_NW as the sum of
wndWNW + wndNW.
This sum of wind direction counts per time
period is due to a statement by Dr. Laurie Goodrich in January
2023 that
observers began using the three-letter
designations such as WNW in 1995, accounting for the drop in NW
wind counts.
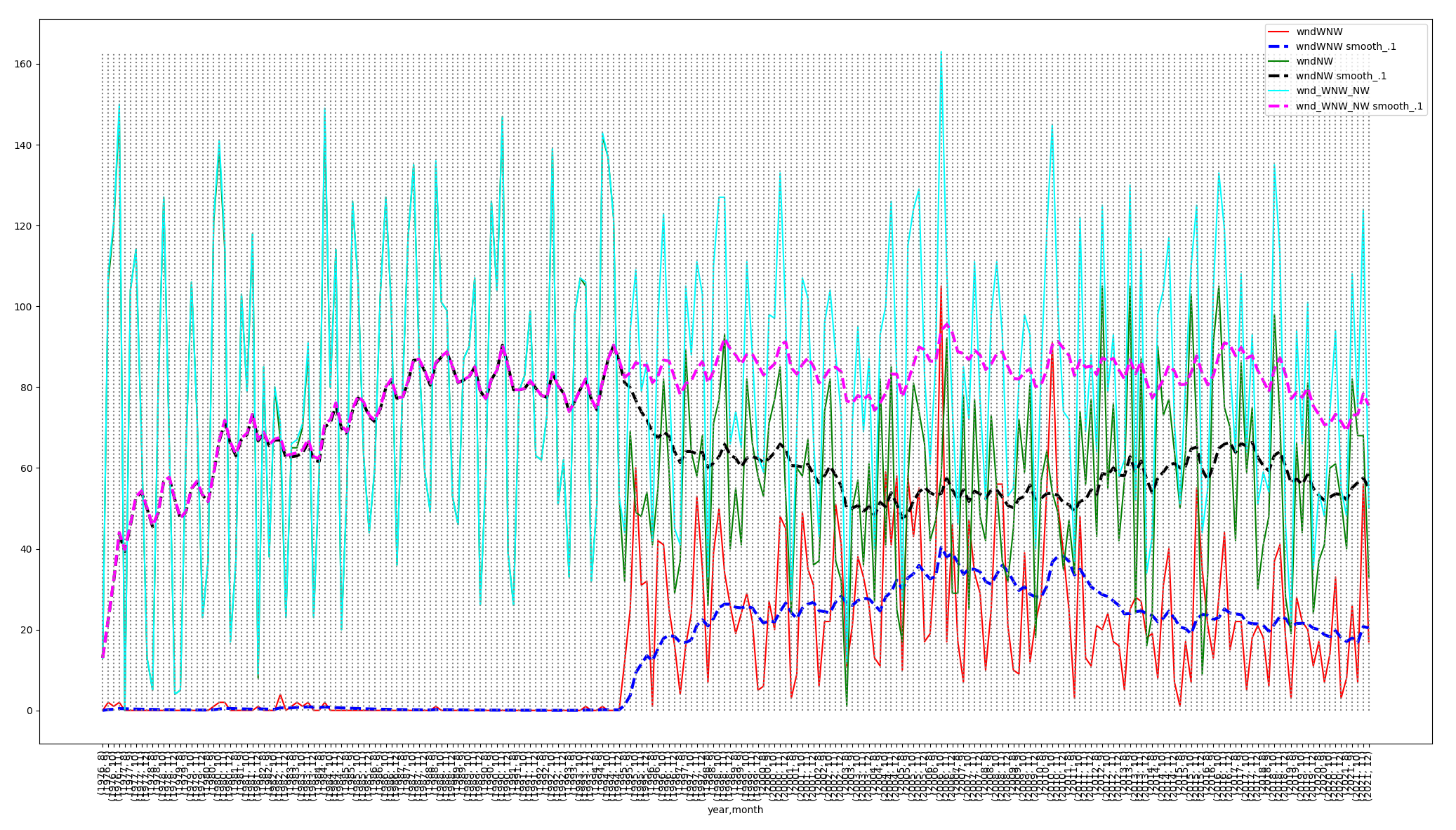
The dashed, exponential smoothing curves in the above graph
compute a weighted estimate at each time step:
Estimatetime = alpha X
Valuetime + (1 - alpha) X Estimatetime-1,
with alpha = 0.1 in this plot.
The plot shows that adding WNW + NW restores the important NW
level from 1995 onward.
Sometimes multi-year averaging can be useful to generate slopes
across multiple years as an alternative to exponential
smoothing.
1c. Remove all raptor attributes containing _All in
their names except for your XX_All attributes.
Keep XX_All, XX_All_log10, and XX_All_td
for your XX in the data.
Remove ONLY the other _All, _All_log10,
and _All_td containing raptor attributes.
1d. Filter -> unsupervised -> attribute ->
reorder the attributes to put your XX_All target
attributes at the bottom of the 60.
Here are my final 10 when XX is SS.
1e. Save this file as XX_month_change.arff.gz, using the arff.gz
format save, where XX is your raptor two-letter ID.
You must turn this into D2L along with XX_week_change.arff.gz in a later step.
Q1: Remove your XX_All_td and XX_All_log10 temporarily and
run the M5P regresssor. Record these results for XX_All as the
target.
(See the README.)
Q2: Temporarily remove year and month because
they are not climate attributes, run M5P again, and record your
results.
(XX_All_td and XX_All_log10 are still out of
the data. See the README.)
Q3: What climate attributes appear in the DECISION TREE part of
Q2's answer. Do not look at the individual LM expressions.
Q4: Execute UNDO until restoring year, month, and all 3 XX_
attributes, or re-load your saved XX_month_change.arff.gz.
Remove
your XX_All_td and XX_All temporarily and run the M5P
regresssor, keeping XX_All_log10;
year & month are in the data.
Record these results for XX_All_log10 as the target. (See the
README.)
Q5: Temporarily remove year
and month because they are not climate
attributes, run M5P again, and record your results.
(XX_All_td and XX_All are still out of
the data. XX_All_log10
is the target. See the
README.)
Q6: Compute the following ratios, where CC is correlation
coefficient, our primary measure of model accuracy.
(CC of Q2) / (CC of Q1) shows drop in model correlation for
XX_All after dropping time and using only climate.
= N.n (the above ratio)
(CC
of Q5) / (CC of Q4) shows drop in model correlation
for XX_All_log10 after dropping time and using only
climate.
Which target approach retains a higher percentage of
CC-with-year-month after it has dropped year and
month,
XX_All or XX_All_log10? Which gives better results
overall in terms of CC, XX_All or XX_All_log10?
Q7: Run SimpleLinearRegression 5 times on the
XX_All_log10 data, with XX_All, XX_All_td, year, and
month OUT of the data.
After each run, record results in the README, then
REMOVE the attribute selected by SimpleLinearRegression
before making
the next test run. You are using the
process of elimination for find the
5 most correlated climate attributes
for SimpleLinearRegression
by finding,
recording, and
removing them, one
at a time. See the
README.
Q8: How do the 5 most important climate attributes selected by
SimpleLinearRegression in Q7 compared with the 5 most important
in Q5? Answer is in term of attribute names, not numbers. See the
README.
Q9: Restore the 5 attributes removed in Q7. We are still
predicting XX_All_log10 with the following out of the data:
year, month, XX_All, and XX_All_td. Run the Select attributes Weka
tab with default hyperparameters. Paste your results
per the README. How do the attributes selected by this step
compare with those of Q5 and Q7 in terms of attribute names?
Q10. Turn XX_month_change.arff.gz into D2L.
*********************************************************
Q11 through Q20: Repeat steps Q1 through Q10 using the same XX as
Q1 through Q10, but this time loading week_change.arff.gz,
analyzing its data, and saving XX_week_change.arff.gz.
One goal here is to determine whether higher resolution in terms
of weekly time periods makes for more accurate or less accurate
CCs than monthly time periods, although there are no questions
about that specfically. Just repeat Q1-Q10 using the weekly
dataset.
The README Q11-Q20 contain the answer templates.
At the end you must turn in your completed README and files
XX_month_change.arff.gz and XX_week_change.arff.gz by the due
date.
