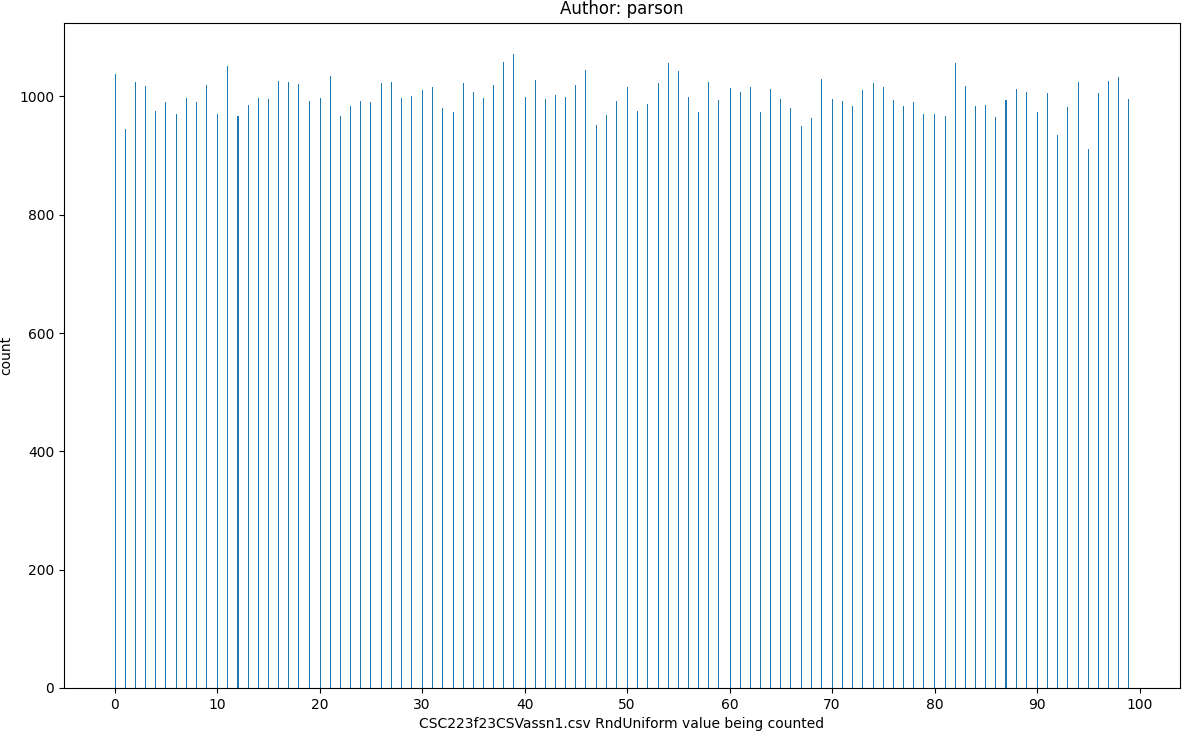
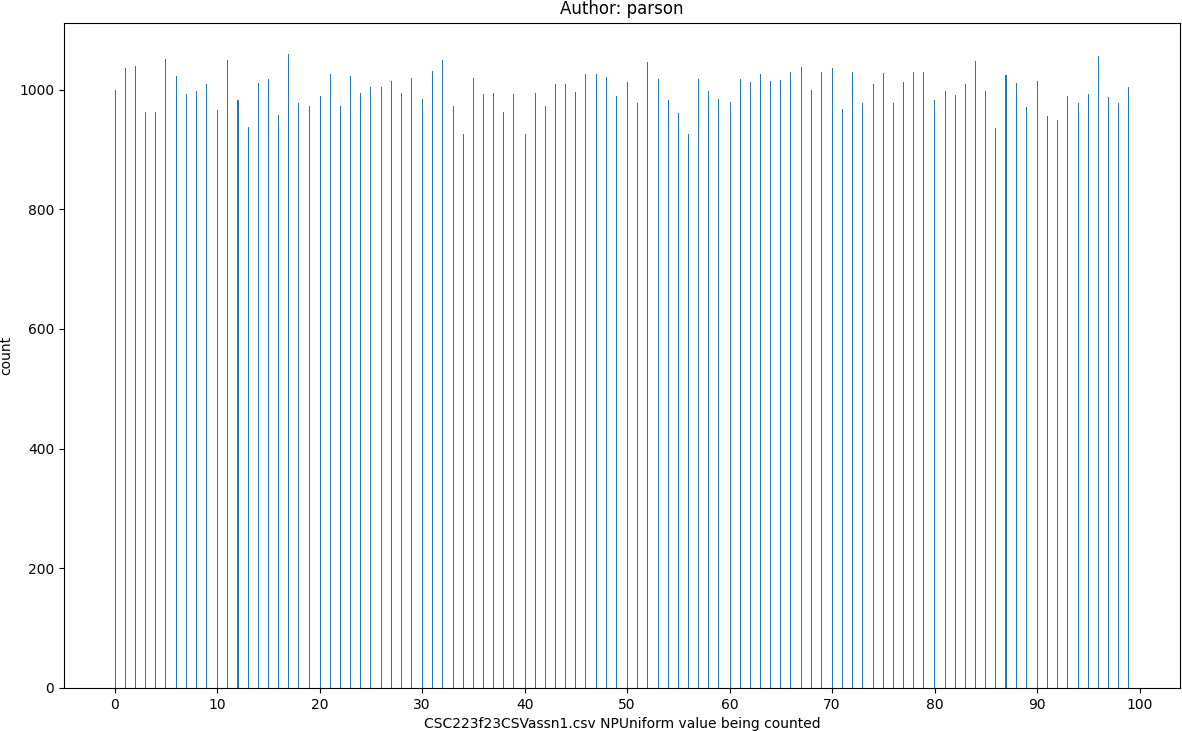
Uniform distribution of 100,000 values in range 0 through 100 from module random

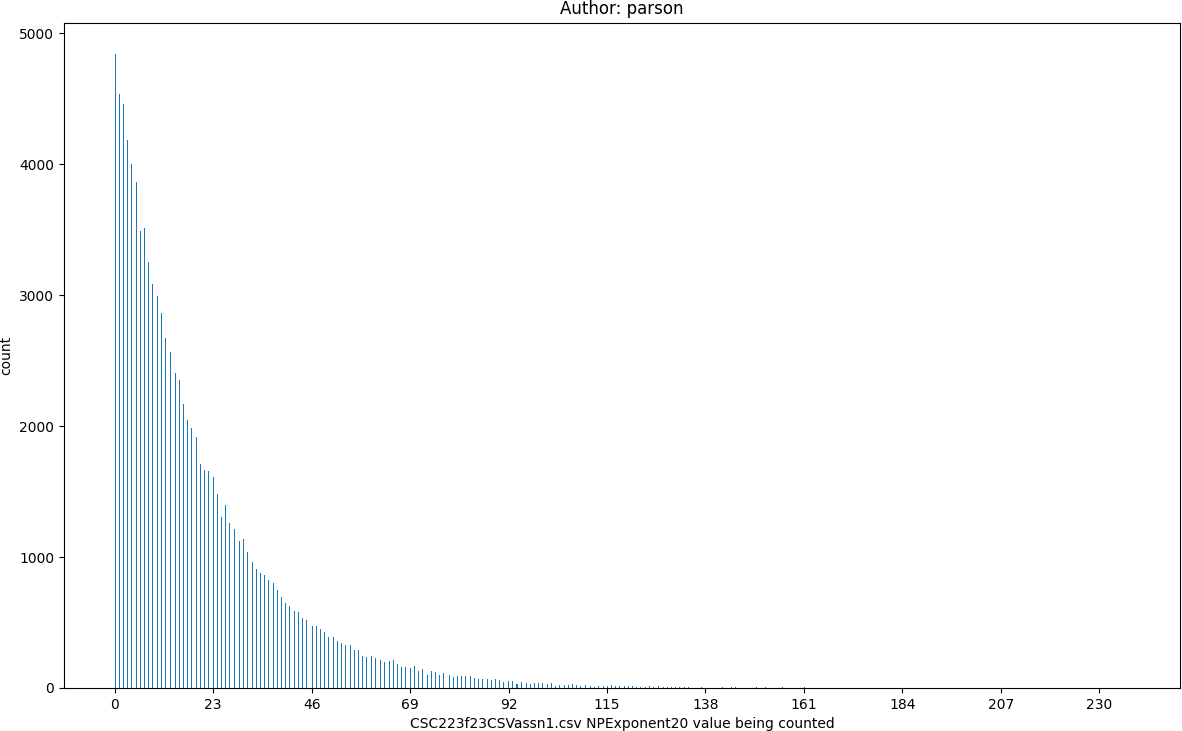
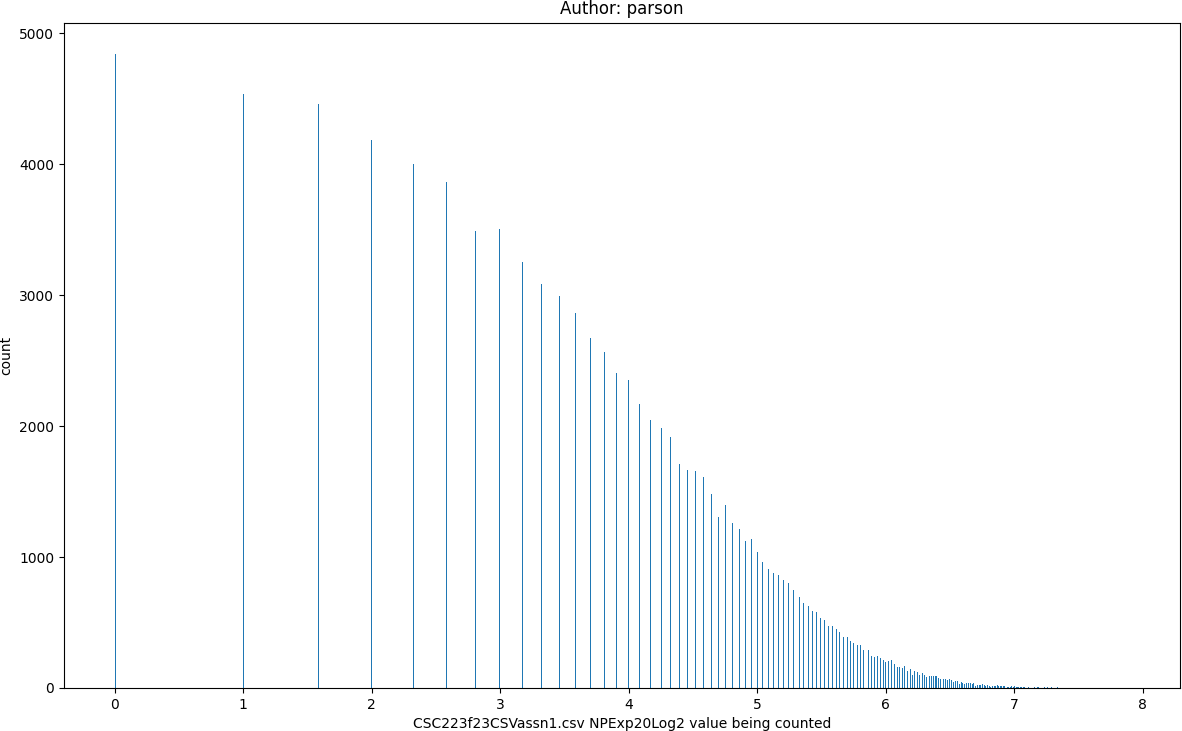
Log2 compresses an exponential range of values into a linear range, which is useful with linear machine learning algorithms
In [8]: log2(0+1) # log of 0 is undefined
Out[8]: 0.0
In [9]: log2(230+1)
Out[9]: 7.851749041416057
Logarithms are reversible.
In [10]: (2**0)-1
Out[10]: 0
In [11]: (2**7.851749041416057)-1
Out[11]: 229.99999999999994
Log2 gives the number of bits in a binary number:
In [12]: values = [2 ** i for i in range(1,11)]
In [13]: for v in values:
...: print(v, log2(v))
...:
2 1.0
4 2.0
8 3.0
16 4.0
32 5.0
64 6.0
128 7.0
256 8.0
512 9.0
1024 10.0
In [19]: from math import ceil
In [20]: values = list(range(2,17))
In [21]: for v in values:
...: print(v, ceil(log2(v)))
...:
2 1
3 2
4 2
5 3
6 3
7 3
8 3
9 4
10 4
11 4
12 4
13 4
14 4
15 4
16 4
$ cat CSC223f23CSVassn1.txt
RndNormal10, seed = 220223523 statistics:
count = 100000
min = 10
max = 93
mean = 49.52
median = 49.0
mode = 49
pstdev = 9.98
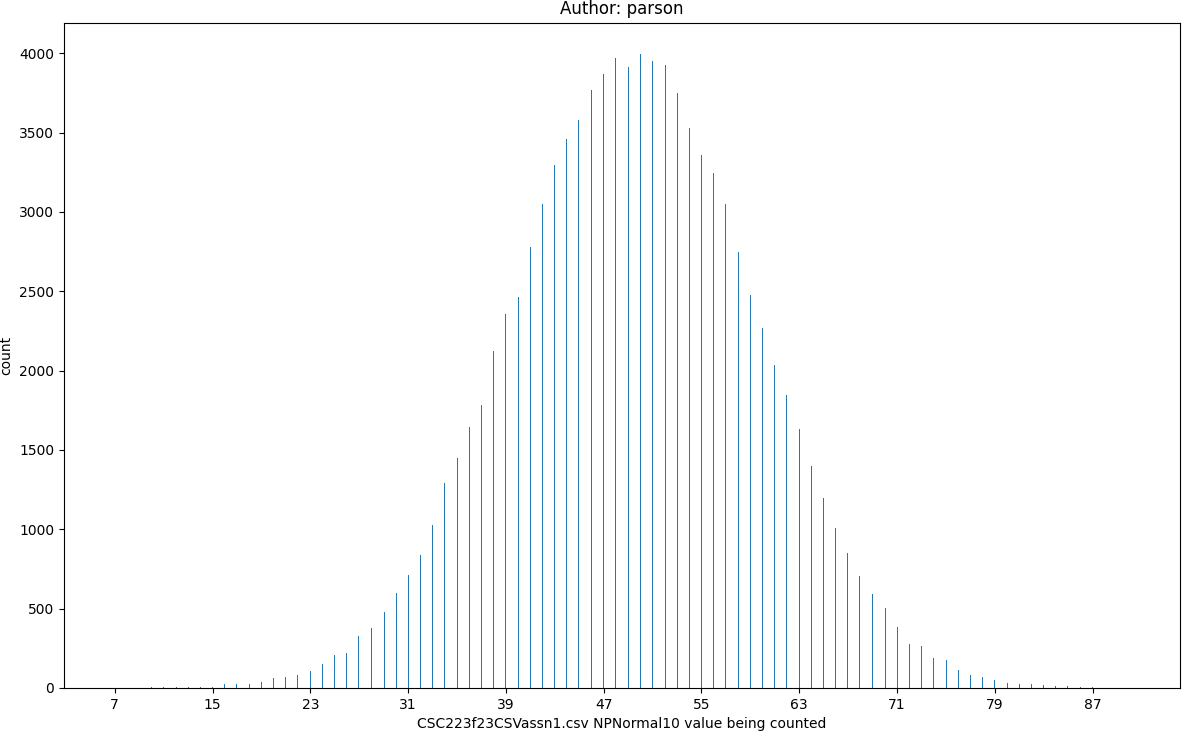
NPNormal10, seed = 220223523 statistics:
count = 100000
min = 7
max = 90
mean = 49.45
median = 49.0
mode = 50
pstdev = 9.99
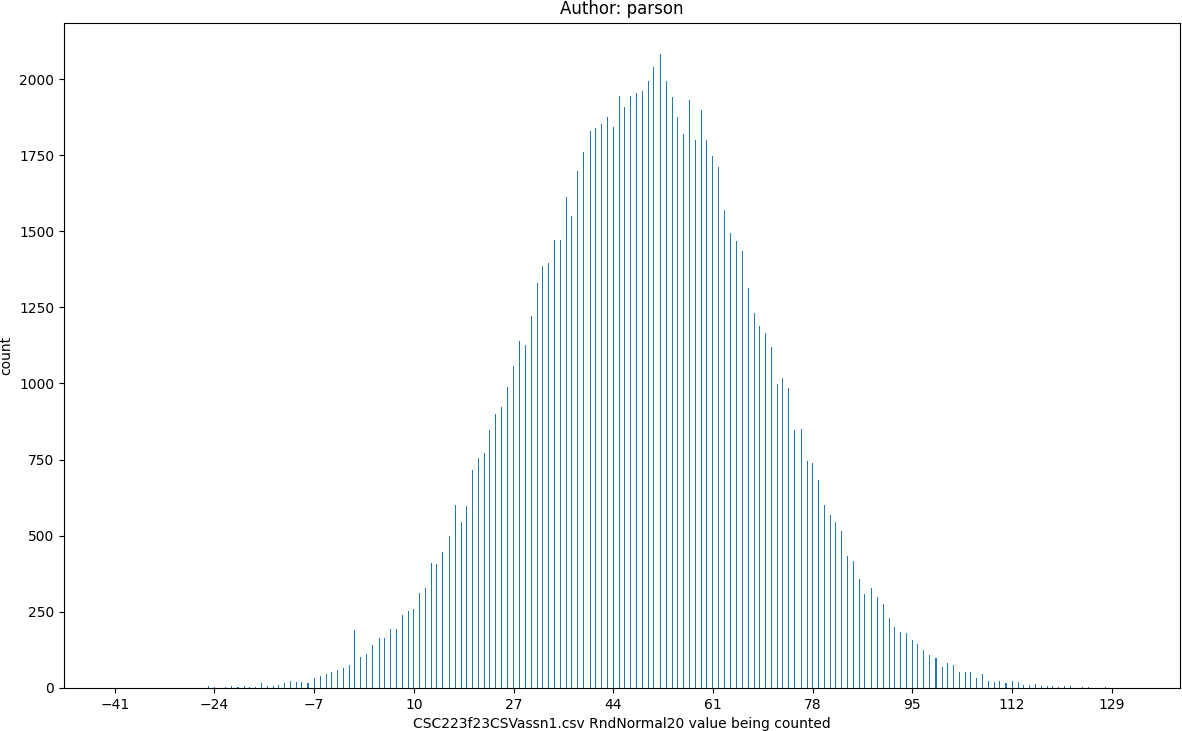
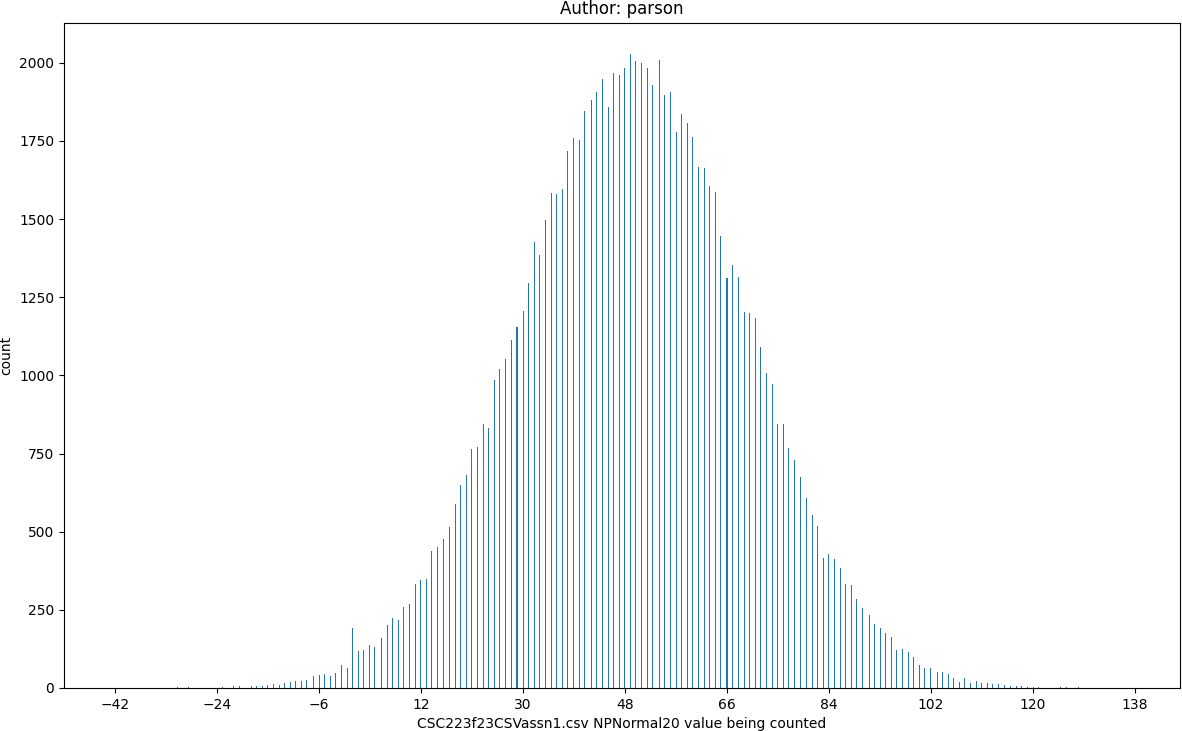
RndNormal20, seed = 220223523 statistics:
count = 100000
min = -41
max = 132
mean = 49.53
median = 50.0
mode = 52
pstdev = 20.05
NPNormal20, seed = 220223523 statistics:
count = 100000
min = -42
max = 137
mean = 49.44
median = 49.0
mode = 49
pstdev = 19.95
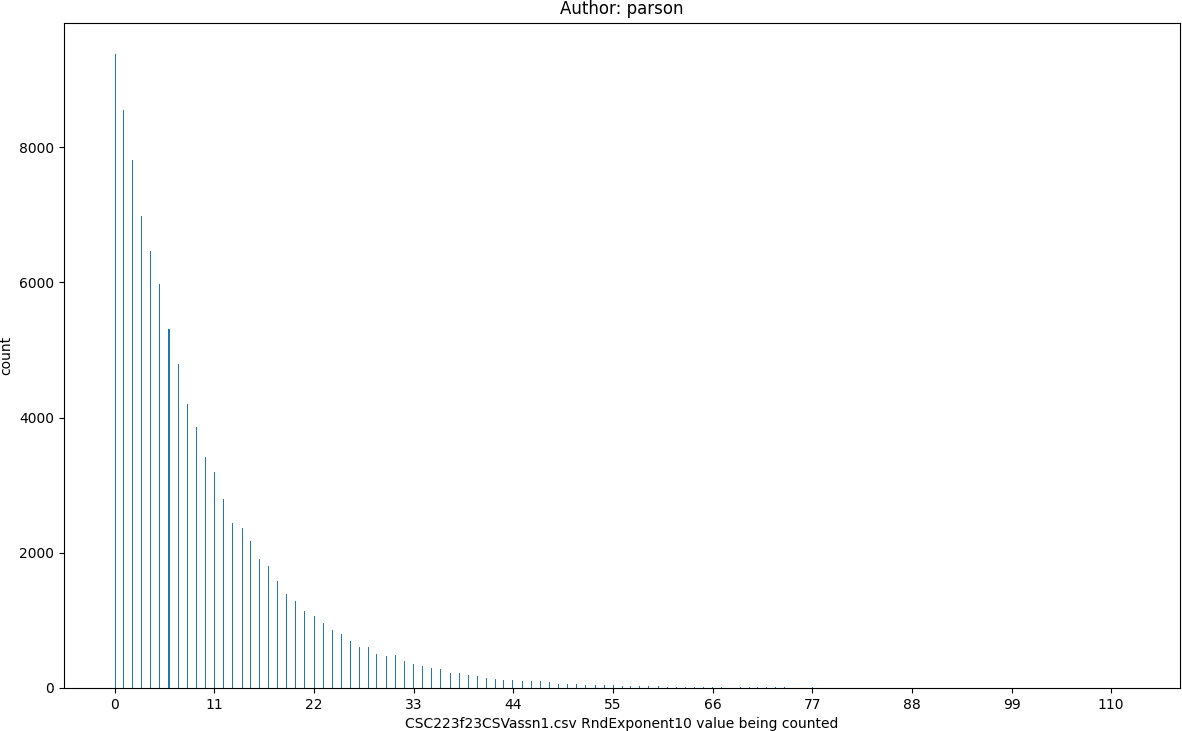
RndExponent10, seed = 220223523 statistics:
count = 100000
min = 0
max = 112
mean = 9.52
median = 6.0
mode = 0
pstdev = 9.99
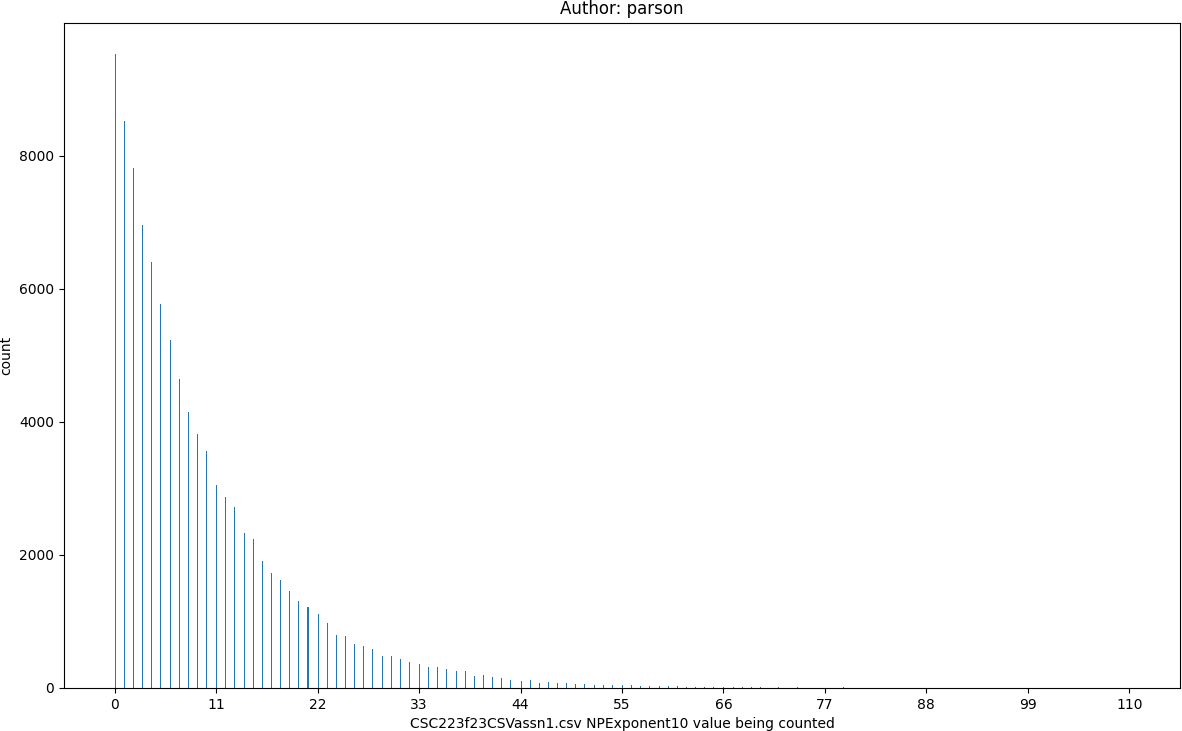
NPExponent10, seed = 220223523 statistics:
count = 100000
min = 0
max = 110
mean = 9.59
median = 6.0
mode = 0
pstdev = 10.1
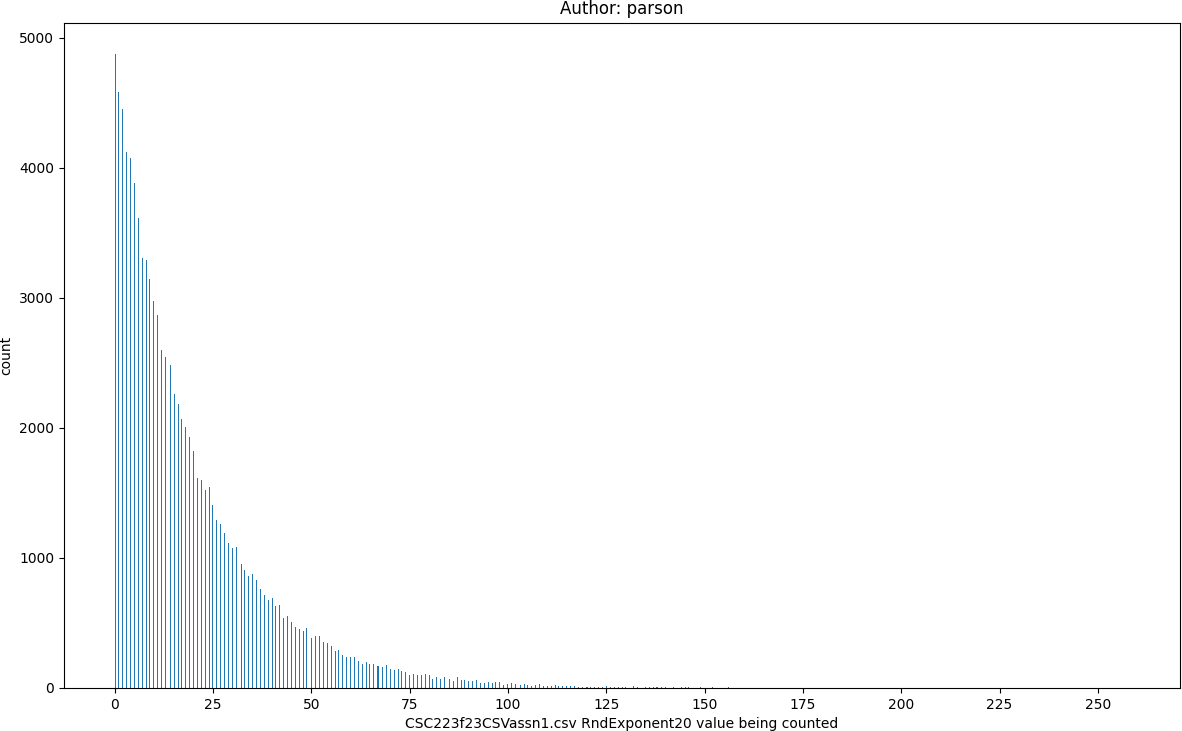
RndExponent20, seed = 220223523 statistics:
count = 100000
min = 0
max = 258
mean = 19.49
median = 13.0
mode = 0
pstdev = 19.96
NPExponent20, seed = 220223523 statistics:
count = 100000
min = 0
max = 237
mean = 19.52
median = 13.0
mode = 0
pstdev = 19.95
NPExp20Log2, seed = 220223523 statistics:
count = 100000
min = 0.0
max = 7.89
mean = 3.65
median = 3.81
mode = 0.0
pstdev = 1.58
From your handout code the following test should work.
$ make clean CSC223f23CSVpre1.csv
/bin/rm -f *.o *.class .jar core *.exe *.obj *.pyc __pycache__/*.pyc
/bin/rm -f junk* *.pyc *.png *.csv CSC223f23CSVpre1.txt
/bin/rm -f *.tmp *.o *.dif *.out __pycache__/* CSC223f23CSVassn1.txt
/bin/rm -f /home/kutztown.edu/parson/tmp/parson*.csv CSC223f23CSVpre1.csv CSC223f23CSVassn1.csv
/bin/rm -f /home/kutztown.edu/parson/tmp/parson*.txt CSC223f23CSVpre1.txt CSC223f23CSVassn1.txt
/bin/rm -f ./CSC223f23CSVpre1.csv ./CSC223f23CSVpre1.txt
/usr/local/bin/python3.7 CSC223f23CSVpre1.py 220223523 /home/kutztown.edu/parson/tmp/parson_CSC223f23CSVpre1.csv
ln -s /home/kutztown.edu/parson/tmp/parson_CSC223f23CSVpre1.csv CSC223f23CSVpre1.csv
ln -s /home/kutztown.edu/parson/tmp/parson_CSC223f23CSVpre1.txt CSC223f23CSVpre1.txt
diff --ignore-trailing-space --strip-trailing-cr CSC223f23CSVpre1.txt /home/kutztown.edu/parson/Scripting/csc223assn1reffiles/CSC223f23CSVpre1.txt > CSC223f23CSVpre1.txt.dif
/usr/local/bin/python3.7 diffcsv.py CSC223f23CSVpre1.csv /home/kutztown.edu/parson/Scripting/csc223assn1reffiles/CSC223f23CSVpre1.csv
FILES CSC223f23CSVpre1.csv,/home/kutztown.edu/parson/Scripting/csc223assn1reffiles/CSC223f23CSVpre1.csv OK.
At that point running make graphs will graph the histograms in any CSV file.
$ make graphs
bash ./makegraphs.sh
mkdir: cannot create directory ‘/home/kutztown.edu/parson/public_html’: File exists
Extracting CSC223f23CSVpre1.csv CSC223f23CSVpre1 RndUniform CSC223f23CSVpre1_RndUniform.png
https://acad.kutztown.edu/~parson/CSC223f23CSVpre1_RndUniform.png
Extracting CSC223f23CSVpre1.csv CSC223f23CSVpre1 NPUniform CSC223f23CSVpre1_NPUniform.png
https://acad.kutztown.edu/~parson/CSC223f23CSVpre1_NPUniform.png
Use your work's graphs for visual detection of bugs and make clobber to remove all PNG files for storage recovery.
$ make clobber
/bin/rm -f *.o *.class .jar core *.exe *.obj *.pyc __pycache__/*.pyc
/bin/rm -f junk* *.pyc *.png *.csv CSC223f23CSVpre1.txt
/bin/rm -f *.tmp *.o *.dif *.out __pycache__/* CSC223f23CSVassn1.txt
/bin/rm -f /home/kutztown.edu/parson/tmp/parson*.csv CSC223f23CSVpre1.csv CSC223f23CSVassn1.csv
/bin/rm -f /home/kutztown.edu/parson/tmp/parson*.txt CSC223f23CSVpre1.txt CSC223f23CSVassn1.txt
/bin/rm -f $HOME/public_html/CSC223f23*.png
If make test fails, look at the non-empty .dif files.
$ ls -l *dif
-rw-r--r--. 1 parson domain users 1543 Sep 9 11:45 CSC223f23CSVassn1.txt.dif
-rw-r--r--. 1 parson domain users 0 Sep 9 11:45 CSC223f23CSVpre1.txt.dif
Here is what a full working make test and looks like.
$ make test
/bin/rm -f *.o *.class .jar core *.exe *.obj *.pyc __pycache__/*.pyc
/bin/rm -f junk* *.pyc *.png *.csv CSC223f23CSVpre1.txt
/bin/rm -f *.tmp *.o *.dif *.out __pycache__/* CSC223f23CSVassn1.txt
/bin/rm -f /home/kutztown.edu/parson/tmp/parson*.csv CSC223f23CSVpre1.csv CSC223f23CSVassn1.csv
/bin/rm -f /home/kutztown.edu/parson/tmp/parson*.txt CSC223f23CSVpre1.txt CSC223f23CSVassn1.txt
/bin/rm -f ./CSC223f23CSVpre1.csv ./CSC223f23CSVpre1.txt
/usr/local/bin/python3.7 CSC223f23CSVpre1.py 220223523 /home/kutztown.edu/parson/tmp/parson_CSC223f23CSVpre1.csv
ln -s /home/kutztown.edu/parson/tmp/parson_CSC223f23CSVpre1.csv CSC223f23CSVpre1.csv
ln -s /home/kutztown.edu/parson/tmp/parson_CSC223f23CSVpre1.txt CSC223f23CSVpre1.txt
diff --ignore-trailing-space --strip-trailing-cr CSC223f23CSVpre1.txt /home/kutztown.edu/parson/Scripting/csc223assn1reffiles/CSC223f23CSVpre1.txt > CSC223f23CSVpre1.txt.dif
/usr/local/bin/python3.7 diffcsv.py CSC223f23CSVpre1.csv /home/kutztown.edu/parson/Scripting/csc223assn1reffiles/CSC223f23CSVpre1.csv
FILES CSC223f23CSVpre1.csv,/home/kutztown.edu/parson/Scripting/csc223assn1reffiles/CSC223f23CSVpre1.csv OK.
/bin/rm -f ./CSC223f23CSVassn1.csv ./CSC223f23CSVassn1.txt
/usr/local/bin/python3.7 CSC223f23CSVassn1.py 220223523 /home/kutztown.edu/parson/tmp/parson_CSC223f23CSVassn1.csv
ln -s /home/kutztown.edu/parson/tmp/parson_CSC223f23CSVassn1.csv ./CSC223f23CSVassn1.csv
ln -s /home/kutztown.edu/parson/tmp/parson_CSC223f23CSVassn1.txt ./CSC223f23CSVassn1.txt
diff --ignore-trailing-space --strip-trailing-cr CSC223f23CSVassn1.txt /home/kutztown.edu/parson/Scripting/csc223assn1reffiles/CSC223f23CSVassn1.txt > CSC223f23CSVassn1.txt.dif
/usr/local/bin/python3.7 diffcsv.py CSC223f23CSVassn1.csv /home/kutztown.edu/parson/Scripting/csc223assn1reffiles/CSC223f23CSVassn1.csv
FILES CSC223f23CSVassn1.csv,/home/kutztown.edu/parson/Scripting/csc223assn1reffiles/CSC223f23CSVassn1.csv OK.
If you want to see histograms for debugging, run make graphs once you have CSV files, then use make clobber to recover space.
$ make graphs
bash ./makegraphs.sh
mkdir: cannot create directory ‘/home/kutztown.edu/parson/public_html’: File exists
Extracting CSC223f23CSVassn1.csv CSC223f23CSVassn1 RndUniform CSC223f23CSVassn1_RndUniform.png
https://acad.kutztown.edu/~parson/CSC223f23CSVassn1_RndUniform.png
Extracting CSC223f23CSVassn1.csv CSC223f23CSVassn1 NPUniform CSC223f23CSVassn1_NPUniform.png
https://acad.kutztown.edu/~parson/CSC223f23CSVassn1_NPUniform.png
Extracting CSC223f23CSVassn1.csv CSC223f23CSVassn1 RndNormal10 CSC223f23CSVassn1_RndNormal10.png
https://acad.kutztown.edu/~parson/CSC223f23CSVassn1_RndNormal10.png
Extracting CSC223f23CSVassn1.csv CSC223f23CSVassn1 NPNormal10 CSC223f23CSVassn1_NPNormal10.png
https://acad.kutztown.edu/~parson/CSC223f23CSVassn1_NPNormal10.png
Extracting CSC223f23CSVassn1.csv CSC223f23CSVassn1 RndNormal20 CSC223f23CSVassn1_RndNormal20.png
https://acad.kutztown.edu/~parson/CSC223f23CSVassn1_RndNormal20.png
Extracting CSC223f23CSVassn1.csv CSC223f23CSVassn1 NPNormal20 CSC223f23CSVassn1_NPNormal20.png
https://acad.kutztown.edu/~parson/CSC223f23CSVassn1_NPNormal20.png
Extracting CSC223f23CSVassn1.csv CSC223f23CSVassn1 RndExponent10 CSC223f23CSVassn1_RndExponent10.png
https://acad.kutztown.edu/~parson/CSC223f23CSVassn1_RndExponent10.png
Extracting CSC223f23CSVassn1.csv CSC223f23CSVassn1 NPExponent10 CSC223f23CSVassn1_NPExponent10.png
https://acad.kutztown.edu/~parson/CSC223f23CSVassn1_NPExponent10.png
Extracting CSC223f23CSVassn1.csv CSC223f23CSVassn1 RndExponent20 CSC223f23CSVassn1_RndExponent20.png
https://acad.kutztown.edu/~parson/CSC223f23CSVassn1_RndExponent20.png
Extracting CSC223f23CSVassn1.csv CSC223f23CSVassn1 NPExponent20 CSC223f23CSVassn1_NPExponent20.png
https://acad.kutztown.edu/~parson/CSC223f23CSVassn1_NPExponent20.png
Extracting CSC223f23CSVassn1.csv CSC223f23CSVassn1 NPExp20Log2 CSC223f23CSVassn1_NPExp20Log2.png
https://acad.kutztown.edu/~parson/CSC223f23CSVassn1_NPExp20Log2.png
Extracting CSC223f23CSVpre1.csv CSC223f23CSVpre1 RndUniform CSC223f23CSVpre1_RndUniform.png
https://acad.kutztown.edu/~parson/CSC223f23CSVpre1_RndUniform.png
Extracting CSC223f23CSVpre1.csv CSC223f23CSVpre1 NPUniform CSC223f23CSVpre1_NPUniform.png
https://acad.kutztown.edu/~parson/CSC223f23CSVpre1_NPUniform.png
Finally, use make turnitin (NOT the turnin scipt) and hit Enter at the prompt. If you make changes after make turnitin,
just run it again to over-write the previous submission. That is due by end of 9/29. I distribute grades via email, not D2L.
$ make turnitin
/bin/rm -f *.o *.class .jar core *.exe *.obj *.pyc __pycache__/*.pyc
/bin/rm -f junk* *.pyc *.png *.csv CSC223f23CSVpre1.txt
/bin/rm -f *.tmp *.o *.dif *.out __pycache__/* CSC223f23CSVassn1.txt
/bin/rm -f /home/kutztown.edu/parson/tmp/parson*.csv CSC223f23CSVpre1.csv CSC223f23CSVassn1.csv
/bin/rm -f /home/kutztown.edu/parson/tmp/parson*.txt CSC223f23CSVpre1.txt CSC223f23CSVassn1.txt
Do you really want to send CSC223f23CSVassn1 to Professor Parson?
Hit Enter to continue, control-C to abort.
/bin/bash -c "cd .. ; /bin/chmod 700 . ; \
/bin/tar cvf ./CSC223f23CSVassn1_parson.tar CSC223f23CSVassn1 ; \
/bin/gzip ./CSC223f23CSVassn1_parson.tar ; \
/bin/chmod 666 ./CSC223f23CSVassn1_parson.tar.gz ; \
/bin/mv ./CSC223f23CSVassn1_parson.tar.gz ~parson/incoming"
CSC223f23CSVassn1/
CSC223f23CSVassn1/makelib
CSC223f23CSVassn1/arfflib_3_3.py
CSC223f23CSVassn1/diffcsv.py
CSC223f23CSVassn1/__pycache__/
CSC223f23CSVassn1/histogram.py
CSC223f23CSVassn1/makegraphs.sh
CSC223f23CSVassn1/CSC223f23CSVpre1.py
CSC223f23CSVassn1/bak/
CSC223f23CSVassn1/bak/CSC223f23CSVassn1.py
CSC223f23CSVassn1/CSC223f23CSVassn1.py
CSC223f23CSVassn1/makefile